Multi-marker approach using C-reactive protein, procalcitonin, neutrophil CD64 index for the prognosis of sepsis in intensive care unit: a retrospective cohort study - BMC Infectious Diseases - BMC Infectious Diseases
Study design and participant enrollment
This was a single-center retrospective study carried out at the First Affiliated Hospital of GuangZhou University of Chinese Medicine. From January 2018 to July 2021, a total of 402 consecutive septic patients (no COVID-19 patients) were admitted to the Intensive Care Unit (ICU) (Fig. 1). The diagnosis was based on Sepsis 3.0 definition. There was no age restriction (except for neonates). The exclusion criteria were as followed: missing date, malignant tumor and immunocompromised state (e.g., long-term use of glucocorticoids, immunosuppressants). To check whether our sample size was large enough to develop a clinical prediction model, we applied the approach proposed by Riley et al. [12]. According to a 28-day mortality of 30% [13] and 5 candidate predictors, at least 323 patients were required. Eventually, 349 patients were included in the study. The survival time of each patient was recorded. If the patient survived more than 28 days, then the length of hospital stay or the time of transfer was recorded.
All the patients included in the study had received antibiotics and then transferred to ICU department. The patients were treated with standard therapeutic strategies under the instructions of Surviving Sepsis Campaign Guideline [1]. All the laboratory tests (including nCD64 index) were routinely performed in our institution for the diagnosis and assessment of disease progression. The characteristics of the study population were summarized in Table 1.
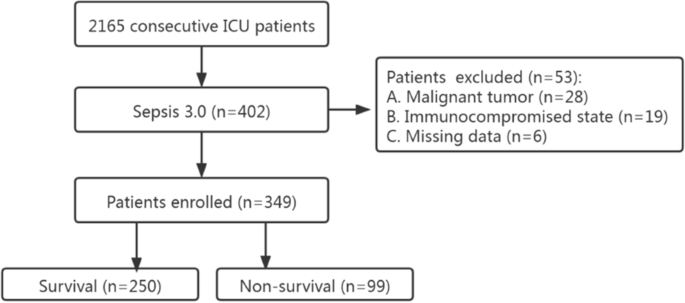
Flowchart of the enrolled patients
Data extraction
Data were retrieved from electronic medical record system in our hospital. Demographic characteristics, histories, comorbidities, site of infection, admission laboratory results, APACHE II and SOFA score were collected. Blood indexes included white blood cells (WBC), neutrophils (NEU), lymphocytes (LYM), platelets (PLT), CRP, PCT, nCD64 index.
Laboratory measurements
Blood samples were drawn from venous puncture right after presentation to ICU and then sent to laboratory department for analysis. CRP levels were quantified by IMMAGE Immunochemistry System (Beckman Coulter, Inc., CA, USA) using nephelometry test. PCT levels were analyzed by serum electrochemiluminescent immunoassay using the mini-VIDAS system (Biomerieux SA, France). The CD64 index was measured using the Cytomics FC 500 MPL (Beckman Coulter, Inc., CA, USA). Specifically, 50 µL of EDTA anticoagulated whole blood was collected and 5 µL CD64-FITC, 5 µL CD33-PE, 5 µL CD45-ECD (Beckman Coulter, Inc., CA, USA) antibody were added to the sample. The sample was then mixed thoroughly and incubated at room temperature in the dark for 15 min. Erythrocyte lysin was then added and mixed and incubated at room temperature in the dark for 10 min. Finally, 500 µL PBS was added. Monocytes, neutrophils and lymphocytes were obtained by CD45/CD33-gating, and the median fluorescence intensity (MFI) of CD64 on the cells was analyzed. nCD64 index = (nCD64 MFI/lymCD64 MFI)/(mCD64 MFI/nCD64 MFI). All the tests were professionally performed by laboratory technicians according to manufacturer's instructions.
Statistical analysis
Data were expressed as median and interquartile range (IQR) or number and percentage. Groups were compared using chi-square test for categorical variables and Mann–Whitney U test for continuous variables. Cox regression model was employed to identify the potential biomarkers for predicting 28-day mortality of sepsis. Hazard ratio (HR) with 95% confidence interval (CI) was utilized for both univariate analysis and multivariate Cox regression model. Age, sex, comorbidities were adjusted for model 1 and those factors plus site of infection, lymphocytes, platelets, APACHE II and SOFA score were adjusted for model 2.
Receiver operating characteristic (ROC) curves was performed to evaluate the predictive accuracy of CRP, PCT, nCD64 index, nCD64 index plus PCT, nCD64 index plus CRP, by comparing the area under curves (AUC). We compared the AUC values among different biomarkers using the method of Hanley and McNeil [14]. Net reclassification improvement (NRI) and the integrated discrimination improvement (IDI) with 95% CI were used to measure the studied models` predictive performance. The optimal cut-off values, sensitivity, specificity, Youden index, positive predictive value (PPV), and negative predictive value (NPV) for each parameter were calculated in predicting the 28-day mortality in septic patients.
All patients were divided into four groups (from 0 to 3) based on the frequency of optimal cut-off values, and each group was compared according to the 28-day mortality using Kaplan–Meier survival curves and HR (with 95% CI). SPSS 23.0 (version 22.0, Chicago, USA), MedCalc Software (version 19.0, MedCalc Software, Mariakerke, Belgium) were used. A two-sided P value < 0.05 was considered statistically significant.

Comments
Post a Comment